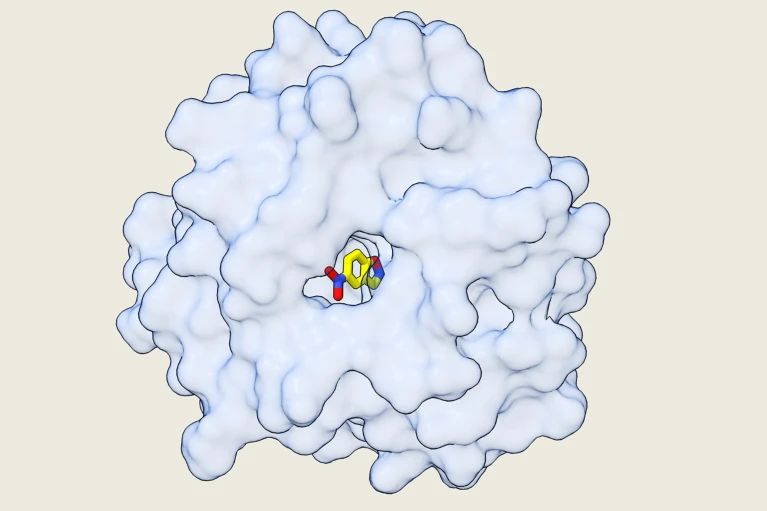
Researchers in synthetic biology have produced the first completely computer-designed enzyme, a synthetic catalyst so effective it surpasses both natural proteins and past AI-generated designs in a historic leap. Published in Nature on June 18, this discovery marks a turning point in protein engineering and opens doors to bespoke enzymes for medicine, green chemistry, and industrial manufacture free from the laborious trial-and-error of conventional lab work.
The Algorithm That Outperformed Evolution

The Kemp elimination is a chemical process that eliminates proton particles from specific carbon-based compounds, and has been identified as the primary reaction of an enzyme that researchers at the Weizmann Institute of Science in Israel seek to create an enzyme that no natural protein could handle. They designed completely new protein structures entirely from scratch, with physics-based algorithms instead of relying on models trained by AI or requiring adjustments to the lab.
The team created thousands of designs possible by separating natural enzymes and reassembling them using computational methods in any possible mix. The team then predicted the structure that could produce functional enzymes that are stable and stable previously unattainable without careful experimentation and tuning.
A Surprising Twist in Enzyme Design
One of the most shocking revelations came when the method disproved a long-held biochemistry theory. Traditionally, scientists thought that the active site of an enzyme in the area where catalysis takes place required a ring-shaped amino acid. But the efficiency of the resulting enzyme shot through when the team’s program recommended a non-ringed amino acid instead.
Lead researcher on the project, Sarel Fleishman, admits he was startled. “These challenges accepted wisdom, but the computer saw what we were unable to see.” Though it was 100 times more efficient than earlier AI-designed models, the final synthetic enzyme differed from its closest natural counterparts by more than 140 amino acids.
Why This Changes Everything for Biotechnology
Designing synthetic enzymes meant beginning with natural proteins and working through expensive, time-consuming lab trials until now. This fresh computing approach turns the script around:
- There is no required natural template. Reactions that nature never evolved for can be created with enzymes.
- More quickly and less expensive development is less dependent on trial-and-error laboratory research.
- Unmatched effectiveness beats both artificial and natural enzymes.
Sílvia Osuna, a computational chemist at the University of Girona who was not involved in the research, notes “they’re remarkable.” “Computationally designing a highly efficient enzyme is quite difficult; this team has done it.”
From Lab Curiosity to Real-World Impact

The implications stretch far beyond academic curiosity. Custom enzymes could revolutionize:
- Medicine Designing enzymes to synthesize drugs more cleanly and efficiently.
- Green Chemistry Replacing toxic industrial processes with enzyme-driven reactions.
- Biofuels Creating optimized enzymes to break down plant matter into energy.
“This is just the beginning,” says Fleishman. “We’re now looking at reactions that were previously unimaginable for enzymes to catalyze.”
The Future: A World of Bespoke Enzymes on Demand?

Should this method scale-up, researchers could soon create enzymes for any chemical reaction on demand. The group is already looking at uses in plastic breakdown, carbon capture, and even new medications.
Still, challenges exist. Although the Kemp elimination enzyme is a proof of concept, modifying this approach for more complicated reactions will need improving the algorithms still more. Still, the success of this entirely computational design points to the arrival of de novo enzyme engineering and its speed surpassing all expectations.
What’s Next?
The researchers plan to expand their library of synthetic enzymes, targeting reactions critical to pharmaceuticals and sustainable manufacturing. Biotech companies, meantime, are closely observing ready to use this discovery for practical purposes.
One thing is clear: the guidelines of protein engineering have recently been revised. And this time the computer performed better than nature.
Sources:

Jan loves Wildlife and Animals and is one of the founders of Animals Around The Globe. He holds an MSc in Finance & Economics and is a passionate PADI Open Water Diver. His favorite animals are Mountain Gorillas, Tigers, and Great White Sharks. He lived in South Africa, Germany, the USA, Ireland, Italy, China, and Australia. Before AATG, Jan worked for Google, Axel Springer, BMW and others.